Contents
- Muse cell therapy for neurological deseases (PI: Niizuma)
- Moyamoya disease and Cerebrovascular disorders (PI: Niizuma)
- Medical Devices development (PI: Niizuma)
- tRNA Biology (PI: Rashad)
- Alternative splicing and RNA binding proteins biology (PI: Rashad)
- Comprehensive image analysis of heterogeneous cells using high-speed imaging technology (PI: Kanno)
Muse cell therapy for neurological deseases (PI: Niizuma)
Stem cell therapy has been a saught after solution for decades to treat many diseases. MUSE (Multilineage stress enduring) stem cells are endogenous pluripotent-like stem cells. They are normally located in the bone marrow, peripheral blood, and connective tissues of organs and are thus non-tumorigenic. MUSE stem cells have been discovered by Professor Mari Dezawa, who is currently a Professor at Tohoku University. In our lab, we carry out extensive work on the utility of MUSE stem cells in treating neurological diseases. Our ongoing work focuses on ischemic and hemorrhagic strokes as well as traumatic brain and spinal cord injuries. Our department spearheaded a clinical study on the use of MUSE cells in ischemic stroke (Niizuma et al, JCBFM, 2023). Our ongoing work focuses on developing novel methods for the preparation and optimization of MUSE stem cells delivery, the expansion of MUSE cells application to various diseases, and understanding the basic biology by which MUSE stem cells offer neuroprotective benefits.
For inquiries regarding this theme, please contact Dr Niizuma: niizuma@nsg.med.tohoku.ac.jp
Moyamoya disease and Cerebrovascular disorders (PI: Niizuma)
Moyamoya disease (MMD) is a rare cerebrovascular disease caused by mutations in RNF213 gene. MMD was discovered by Professor Jiro Suzuki, the first professor of Neurosurgery at Tohoku University in the 1960s. Since Professor Suzuki’s discovery, Tohoku University has been a leader in developing clinical and research tools and methods for treating and understanding MMD. In our lab, we focus on the pathophysiology of MMD from the standpoint of gene mutations in RNF213. We was various tools such as iPSCs drived cells from MMD patients, animal modeling, gene editing, and sequencing technologies to advance our knowledge on MMD pathology and RNF213 gene function. In the past years, our lab have developed many novel concepts in MMD research, such as the role of RNF213 in immunity and the links between RNF213 and post-transcriptional mRNA splicing. We continue to expand our work on MMD pathology with a special interest on using iPSCs derived cell modeling and many novel tools and techniques.
For inquiries regarding this theme, please contact Dr Niizuma: niizuma@nsg.med.tohoku.ac.jp
Medical Devices development (PI: Niizuma)
under construction
tRNA Biology (PI: Rashad)
The tRNA epitranscriptome has emerged as important regulator of mRNA translation to proteins, and its role was shown to impact many diseases. Our lab is pioneering is studying many aspects of tRNA biology in oxidative stress and diseases such as tRNA cleavage and tRNA modifications. We currently use state-of-the art tools such as high throughput liquid chromatography tandem mass spectrometry (LC-MS/MS), sequencing technologies, and gene editing to study tRNA biology at the basic and translational levels. Our ongoing projects focus on oxidative and mitochondrial stresses, glioma, diabetes, neurodegenerative diseases, stroke, and neuronal differentiation. In addition, we collaborate with other scientists and researchers to study tRNA biology in other non-neurological conditions such as various cancers, osteoporosis, and more. We also have a special interest in developing novel gene therapies and mRNA vaccine optimization by using principles of codon optimization based on our knowledge of tRNA biology.
Our aim in this field is to develop novel therapies and biomarkers for diseases as well as developing the next generation gene/mRNA therapeutics. In addition, we have special interest in understanding basic tRNA biology and continue to develop tools and ideas to achieve that.
For inquiries regarding this theme, please contact Dr Rashad: sherif.mohamed.rashad.e3@tohoku.ac.jp
Alternative splicing and RNA binding proteins biology (PI: Rashad)
Alternative splicing is a process by which different mRNA isoforms can be produced from a single mRNA. This process is regulated by many factors, such as mRNA modifications and RNA binding proteins (RBPs). In our research, we focus on the role of RBPs in regulating many physiologic and pathologic processes via altering mRNA splicing. Our current interests include, and not limited to, neuronal differentiation, cellular stress, and cancer biology (in particular glioma pathophysiology). In this work, we use various sequencing tools and computational methods to study alternative splicing transcriptome-wide and identify RBPs regulators.
For inquiries regarding this theme, please contact Dr Rashad: sherif.mohamed.rashad.e3@tohoku.ac.jp
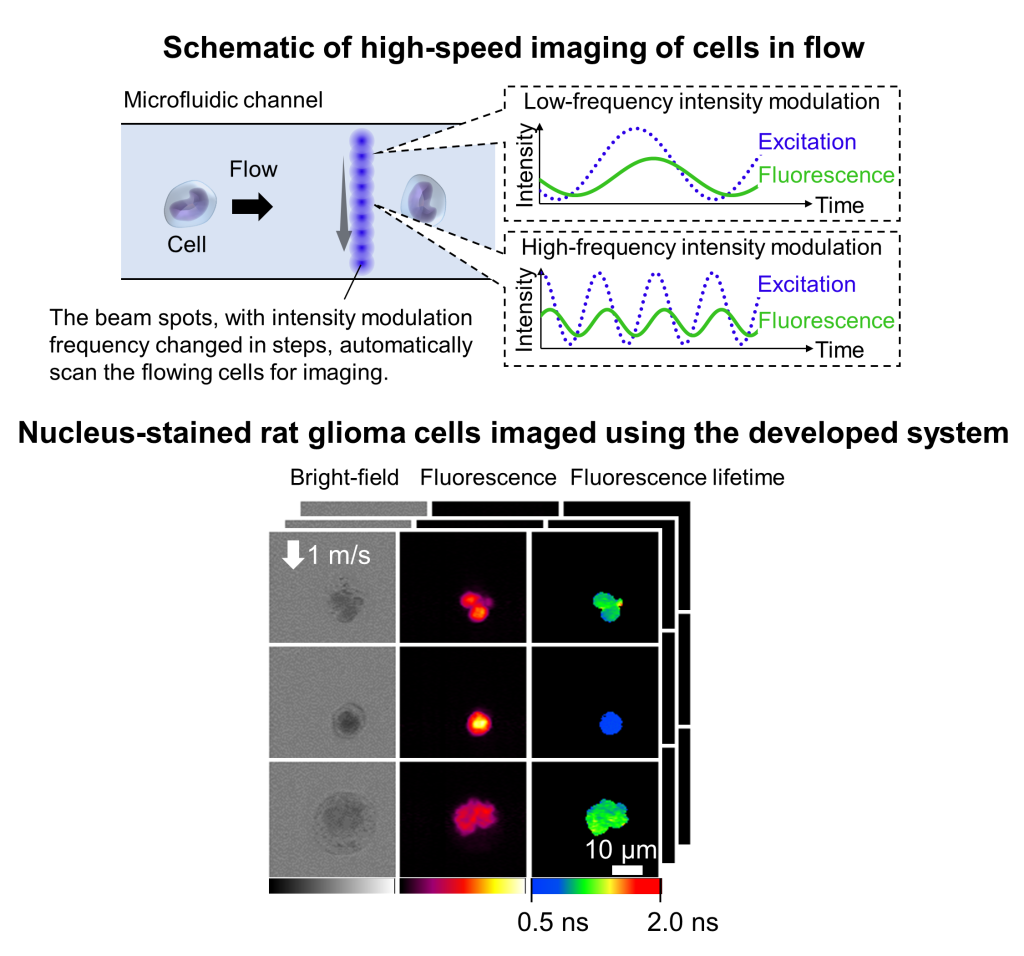
Comprehensive image analysis of heterogeneous cells using high-speed imaging technology (PI: Kanno)
Biological tissues, such as tumors and blood, are composed of cells with diverse types, properties, and states. Therefore, to gain a deeper understanding of the characteristics of these tissues, it is essential to comprehensively analyze the heterogeneous populations of cells that constitute them. However, conventional techniques for cellular analysis, such as microscopy and flow cytometry, fall short in providing detailed, statistically significant analyses of these diverse cell populations.
In our laboratory, we are developing high-speed imaging technologies to facilitate the comprehensive image analysis of heterogeneous cell populations. Specifically, as illustrated in the figure below, we align beam spots in a row within a microfluidic channel and simultaneously measure multiple spatial points, enabling rapid imaging of flowing single cells (>10,000 cells/sec). Each beam spot is intensity-modulated at different frequencies, allowing the signals to be separated and reconstructed through signal processing techniques.
To date, we have applied this technology to the analysis of blood and tumor cells, contributing to studies such as those related to thrombosis in COVID-19 (M. Nishikawa, H. Kanno, Y. Zhou et al., Nature Communications 2021). In addition, we have recently developed the world’s fastest fluorescence lifetime imaging microscopy (FLIM), further enhancing the imaging capabilities for cellular analysis and advancing biomedical applications (H. Kanno et al., Nature Communications 2024). Please refer to the press release for more information on the high-speed FLIM.